web
page
web
page
1. inTag purpose
inTag is a software that facilitates the processing of Tagged MR Image sequences. One of the main reasons of such images not to be used in clinical practice is the lack of appropriate processing tools. inTag offers a solution for the complete and rational analysis of such sequences and results in cartographies, graphs and documented files for easy exploitation by the clinician or scientist.
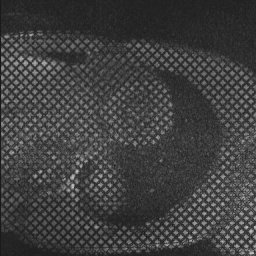
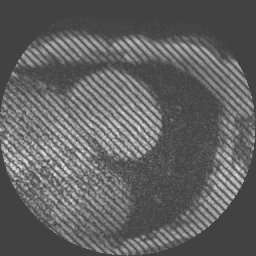
Examples of such sequences with a grid tag and stripe patterns can be seen below:
inTag can process Dicom tagged MRI mono or multi-slice sequences with limited users’ intervention. It extracts a motion field in the entire image or in a Region of Interest (ROI) and the contours of the left ventricle. Finally, it generates color maps, motion fields, graphs and files of diverse motion parameters: motion magnitude, principal deformations, circumferential and radial deformations through the cardiac cycle. The movies and displays below illustrate the kind of representations generated using inTag.
|
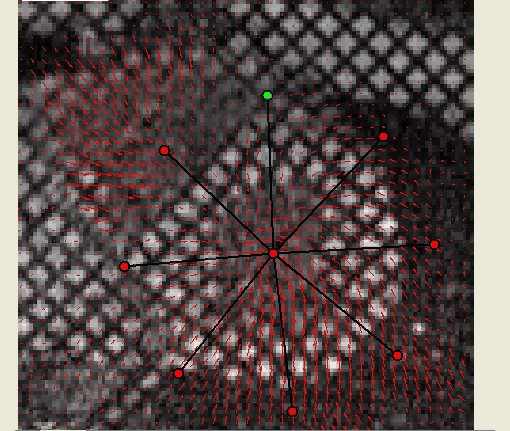
Motion Field of the grid tagged sequence above |
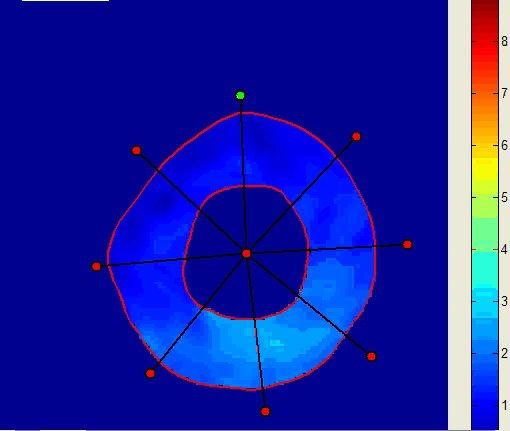
Motion magnitude |
|
|
|
|
Maximum principal strain similar to |
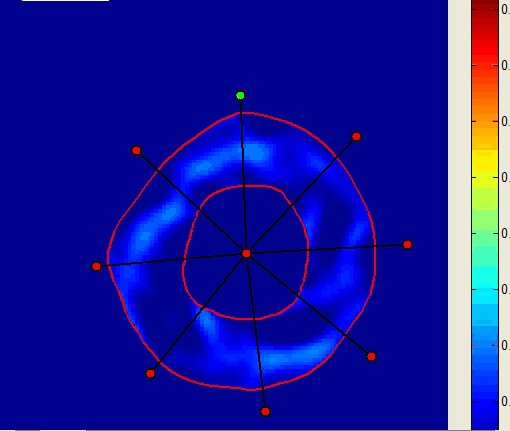
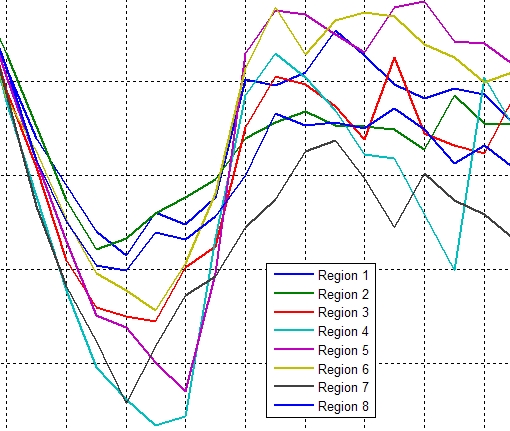
Graphs of the minimum principal strain value in the 8 sectors |
Note that the results are obtained with very few users’ inputs at the very beginning of the process: specifying the tagging pattern, its orientation, the approximate end-diastolic time point and a ROI. The number of sectors, layers and interventricular reference point are specified during the course of the process.
1. inTag process
The processing of tagged MRI sequences is sequential. At the beginning, you have to prepare the images to be interpreted by inTag and then, run inTag. You first indicate the directory where the image files are and specify the tagging pattern, its orientation, the approximate end-diastolic time point and a ROI. inTag will then compute the motion throughout the temporal sequence and the slice levels. The left ventricular myocardium segmentation starts from an initial ring that will be adapted to the heart’s border. The user has just to validate the initialization proposed by the software and then the program proceeds to the last step to produce results like the ones presented above.
A detailed users’ guide is available as the pdf file inTagUsersGuide.pdf.
An example of the processing of an image series is given in this movie (short axis) or movie (long axis).
2. Downloading inTag
To use inTag for PC running Windows, please follow the steps below:
· Retrieve and fill in the users agreement document for academic institutions:
§ The users agreement in French Accord_de_pret_Logiciel_ inTag_CreatisCNRS.doc
§ The users agreement in English Accord_de_pret_Logiciel_ inTag_CreatisCNRS.doc
Everything’s prepared. You just have to read and complete your institution information, give a short description of the context within which you will use inTag and put authorized signatures.
· Send the document by regular mail to the following address:
À L’attention de Patrick Clarysse
Creatis UMR CNRS 5515, Inserm U630
INSA Bâtiment Blaise Pascal
7, Avenue Jean Capelle
69621 Villeurbanne Cedex
France
· Ask for a password by sending a mail to: mailto:patrick.clarysse@creatis.insa-lyon.fr
· To retrieve the automatic installer thanks to the given password for inTag v1.1 InTag_Setup_1.1.exe
· To retrieve the automatic installer thanks to the given password for inTag v1.2 InTag_Setup_1.2.exe
· To retrieve the automatic installer thanks to the given password for inTag v1.3 InTag_Setup_1.3.exe NEW (April 10, 2007)!
· Run inTag to check for the installationwith thesample image sequence.
Note: The code is given as is. Creatis cannot insure a permanent maintenance of the software. You can communicate about the encountered bugs or make suggestions through inTag mailing list (see below). Bugs and software evolution will be processed with no guarantee. In any case, Creatis cannot be considered as responsible for any misinterpretation of the results or software dysfunctions.
3. inTag Mailing list
There exists a mailing list of inTag users. Its address is : mailto:intag@creatis.insa-lyon.fr. To register to this mailing list, please go to http://www.creatis.insa-lyon.fr/mailman/listinfo/intag.
You can ask questions about the software, report for bugs and be aware of corrected inTag versions.