Carole Lartizien
Objectives
Our first objective is to develop and evaluate Monte Carlo simulations for 3D Positron Emission Tomography (PET).
Simulated data are of foremost importance for the design of new tomographs and the development of new PET imaging processing methods. Some Monte Carlo simulation tools are available through the international community, each of them compromising some advantages and drawbacks. Our goal is to promote the use of these simulation tools by allowing realistic simulation of accurate biological radioactive distribution in a reasonable computation time. We now focus on two simulation tools:
- PET-SORTEO (http://sorteo.cermep.fr/)
- GATE based on the Geant4 library (http://opengatecollaboration.org/).
Our second objective is to generate 3D and 4D PET databases of human and small animal images.
Results
-
Development and validation of Monte Carlo simulation tools
PET-SORTEO was validated against the geometries of the two widely distributed microPET R4 and Focus 220 systems manufactured by Siemens Preclinical Solutions
Figure 1. Projection images of a 300 s acquisition of the MOBY phantom with 185 kBq (∼5 µCi) in the mouse bones on the R4 (left) and Focus 220 (right) systems.
The reconstructed pixel size is 0.84 × 0.84 × 1.21 mm3 (in x, y and z directions) with the R4 system and 0.95 × 0.95 × 0.796 mm3 with the Focus system.
-
Construction of a realistic 3D PET whole-body image database
Our aim was to generate and distribute a realistic database of simulated whole body [18F]FDG PET oncology images. OncoPET_DB (http://www.creatis.insa-lyon.fr/oncoPET_DB) provides 100 realistic simulated [18F]FDG PET images with (50 cases) or without (50 cases) inserted lesions. As far as we know, this database is the first addressing the need of simulated [18F]FDG PET oncology images by providing a series of whole-body (WB) patient images with well controlled inserted lesions of calibrated uptakes. It also will fulfil the requirements of detection performance studies by including normal and pathological cases. The realism of the database is based on three points: firstly, we built a complex model of [18F]FDG patient based on the Zubal phantom in combination with activity distributions in the main organs of interest derived from a series of 70 clinical cases. Secondly, we proposed a realistic model of lesions extent corresponding to lymphoma patients. Finally, the simulated database was generated with the PET-SORTEO Monte-Carlo simulation tool that was fully validated against the geometry of the ECAT EXACT HR+ (CTI/Siemens Knoxville).
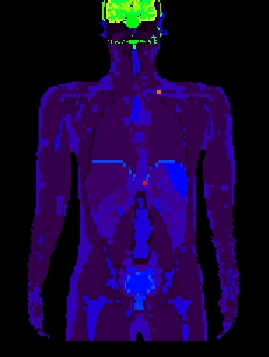
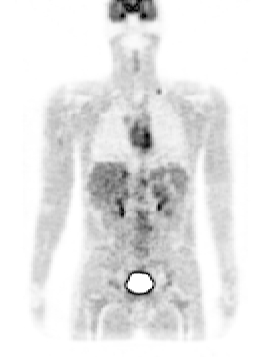
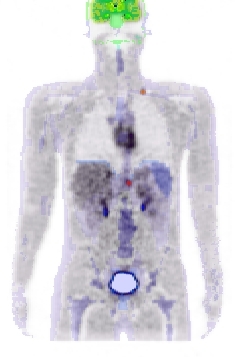
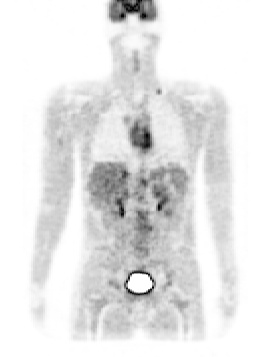
Figure 2. left: coronal slice through the Zubal phantom with inserted lesions (red points). Middle: simulated PET image corresponding to the Zubal model. Right: superimposed label model and PET simulated data.
Perspectives include enriching the oncoPET_DB with new cases based on the breathing NCAT model to account for respiratory motion and anatomical variability.