Demo session at the Interactive Medical Image Computing (IMIC) Workshop - MICCAI 2014.
Overview
inTag, CMRSegTools and C-DTI are a set of plug-ings for the OsiriX DICOM viewer. These tools have been designed to exploit OsiriX intuitive navigation and interactive visualisation functionalities for the analysis of cardiac MR images. Each plug-ing implements a well-defined workflow and novel interaction techniques in order to simplify and streamline analysis tasks.
inTag
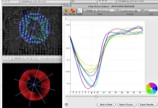
strains plots.
inTag has been developed in order to compute and interact with myocardial strain analysis from tagged MR sequences. The user is guided by the program into five steps: specify input data settings (i.e. tag orientation, tag spacing), select a region of interest (ROI), compute motion fields (based on the SinMod method [ARTS-10]), compute the endocardium and epicardium contours (based on the dynamic deformable elastic template [SCHA-10]), compute and display analysis strain graphs (i.e. principal strains, circumferential and radial deformations, among others).In the next inTag release, the user may interactively adjust myocardium contours for better results and also may overlap reference graphs (i.e. normal strain values), in order to compare and analyse its current study against statistical information. The statistical references are being made based on contributions of the inTag users community.
CMRSegTools
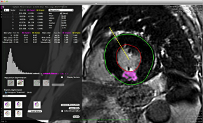
CMRSegTools is designed to ease cardiac and region segmentation of delayed-enhancement images for infarct sizing but also of parametric maps (T1, T2). As a first step, the user is assisted by the program to define the myocardium and compute the endocardium and epicardium contours in 2D or 3D (based on B-spline explicit active surfaces [BARB-12a]). Then, interactive segmentation methods are available: standard mean ± xSD, 2D- or 3D- full-width at half-maximum (FWHM) or more advanced such as Gaussian Mixture Model (GMM), and Hsu method.
C-DTI
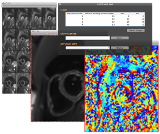
C-DTI is the most recent plug-in implemented to assist users in the extraction of information from cardiac DTI data: mean diffusivity and diffusion anisotropy indices, etc.
References
[SCHA-10] J. Schaerer, C. Casta, J. Pousin, and P. Clarysse, "A dynamic elastic model for segmentation and tracking of the heart in MR image sequences", Medical Image Analysis, vol. 14, no. 6, pp. 738-749, 2010.
[ARTS-10] T. Arts, F. W. Prinzen, T. Delhaas, J. R. Milles, A. C. Rossi, and P. Clarysse, "Mapping Displacement and Deformation of the Heart With Local Sine-Wave Modeling", IEEE Transactions on Medical Imaging, vol. 29, no. 5, pp. 1114-1123, 2010.
[BARB-12a] Barbosa, D., Dietenbeck, T., Schaerer, J., D'hooge, J., Friboulet, D., Bernard, O., "B-Spline Explicit Active Surfaces: An Efficient Framework for Real-Time 3-D Region-Based Segmentation", Image Processing, IEEE Transactions on , vol.21, no.1, pp.241,251, Jan. 2012.
R&D Team at CREATIS
| William A. ROMERO R. |
Research Engineer.
|
| Pierre CROISILLE | Chairman Department of Radiology at CHU Saint-Etienne - Université Jean-Monnet. Deputy Director at CREATIS. |
| Magalie VIALLON |
MR physicist at Hopital Universitaire de Genève, University of Geneva and CHU Saint-Etienne.
|
| Patrick CLARYSSE | Research Scientist - Centre national de la recherche scientifique (CNRS). Leader of the team 1 at CREATIS. |
| Yuemin ZHU |
Research Scientist - Centre national de la recherche scientifique (CNRS).
|








