Name of the coordinator (team): Maciej ORKISZ (MYRIAD)
Names of permanent staff involved: Odyssée MERVEILLE (MYRIAD), Emmanuel ROUX (MYRIAD), Olivier BERNARD (MYRIAD), David SARRUT (TOMORADIO), Thomas BAUDIER (TOMORADIO), Thomas GRENIER (MYRIAD), Jean-Christophe RICHARD (MYRIAD), Carole LARTIZIEN (MYRIAD), Carole FRINDEL (MYRIAD), Benjamin LEPORQ (MAGICS), Pierre CROISILLE (MAGICS), Jean-Baptiste PIALAT (MAGICS), Frederic Cervenansky (INFO-DEV), Eduardo DÁVILA (INFO-DEV), François COTTON (MAGICS)
Context
The project described below was initiated at the beginning of the COVID-19 pandemic and was active until the end of 2023. It progressively evolved towards a smaller group focusing on Functional Imaging and Lung Modeling (see FILM project). Main results achieved are listed at the end.
Since the beginning of the COVID-19 pandemic, radiologists and intensive-care doctors, members of CREATIS examine patients for diagnostic and/or therapeutic-choice purposes. With help of CREATIS IT engineers, two rich databases were constructed and continue growing, which include pulmonary CT images as well as very rich complementary patient data. CREATIS researchers joined their efforts to exploit these data through the development of efficient image processing tools, in order to contribute to the knowledge of the disease. In the meanwhile, neuroradiologists of the lab, under the initiative of the French Neuroradiological Society (SFNR) contributed to the delineation of a large cohort of confirmed brain MRI parenchymal signal abnormalities (excluding ischemic infarcts) in patients with severe COVID-19. This multicentric database (16 clinical centers) provides a first description of the neuroimaging findings (excluding ischemic infarcts) as well as the clinico-biological profile of these patients. CREATIS researchers and neuroradiologists have been since discussing clinical questions regarding the neurological impact of non-severe and severe COVID-19 patients. This opened the way to the definition of methodological developments that will allow the efficient extraction of information from these neuroimaging data to help understand the underlying pathophysiological mechanisms of this disease. Finally, IT engineers from the info-dev team of CREATIS have played an active role during the pandemic period to build, store and provide a secured access to the pulmonary databases. One of these databases is being made available to the French scientific community via secured access. The objective is now to set up a platform that will allow a secured access and the deployment of learning algorithms from an innovative database.
This project is conducted in association with the COVID-19 initiative of CNRS GdR ISIS.
Goal
The overall goal of this project is to develop image-processing methods (mainly based on artificial intelligence) for the diagnosis and prognosis of COVID-19 as well as the understanding of the underlying physiopathological mechanisms. The objective regarding the pulmonary application is to take an additional step in prediction, in particular to detect severe COVID-19 patients at an early stage to help intensive-care unit capacity management. Regarding the neurological application, the main goal is to characterize the brain patterns of the pathology and their correlation to neurological sequelae, especially in non-severe COVID-19 patients.
Three work packages were defined to explore different avenues.
WP 1: Lung and thorax CT analysis
(leaders D. Sarrut and O. Merveille)
- CTVI (computed-tomography-based ventilation imaging): the goal is to develop robust methods capable of calculating 3D ventilation maps from CT image pairs (inhale-exhale) and determine whether or not the disease evolution can be correlated with specific ventilation patterns.
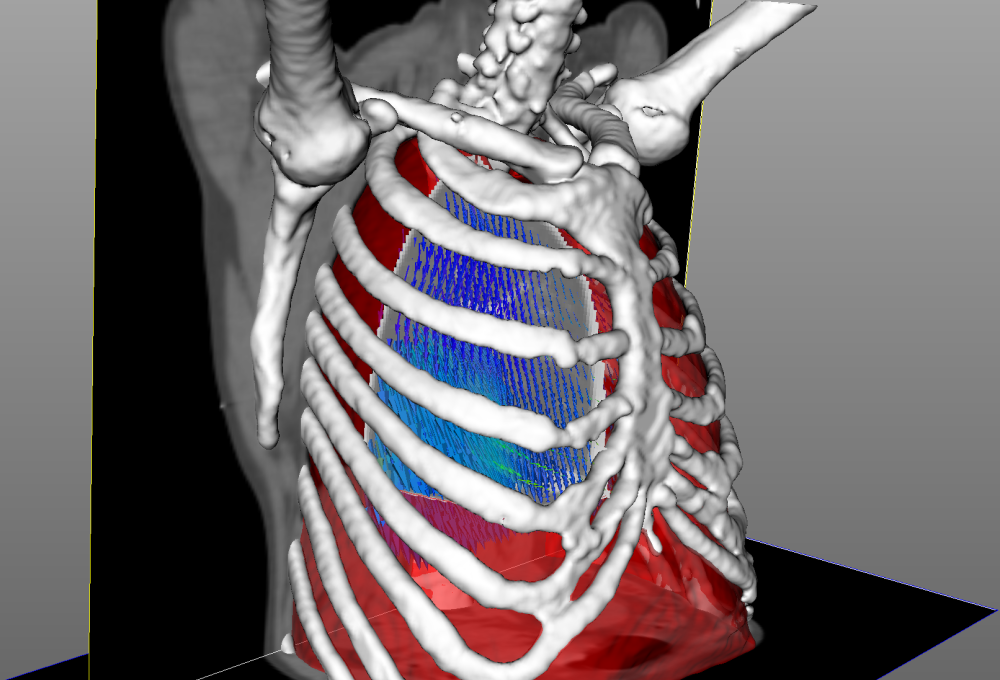
- Lung segmentation: quantifying pathologic changes requires lung delineation, as well as aligning the pulmonary anatomic structures by image registration, the latter using so-called motion masks separating inner moving tissues from the rib-cage, etc.; the goal is to develop segmentation methods robust to locally decreased (or lacking in severe cases) contrasts due to pathologic consolidations.
- Vascular-tree segmentation: the goal is two-fold. First, automatically extract the pulmonary vascular tree in order to help the clinicians study the hypothesis of a change in the vessel diameter distribution induced by COVID-19. Second, detect pulmonary embolism and localize the emboli in the pulmonary vascular tree, which is an important information both for treatment and research purposes.
- Spinal bone and muscle segmentation: the goal is to provide tools for the study of associations between the disease and sarcopenia. This sub-project, SARCOV-IA with the HCL, is an ancillary study focussing on skeletal muscle, fat, and bone assessment on thoracic CT scanner to determine the sarcopenic, osteosarcopenic, and sarcopenic obesity status of the patients. Segmentation will be performed using semi-automatic thresholding with manual adjustment, and then a deep network will be trained to develop automatic segmentation of muscle, fat, and bone on sarcopenia’s specific CT slices. The hypothesis to test is increased mortality in ICU of the patients sustaining one of these pathologic conditions.
- Radiomics analysis to predict the outcomes of the disease. Lung segmentation and registration is a crucial step, since it is mandatory to extract radiomics features. Radiomics features could be next integrated with other data such as clinical data.
Expected results
- CTVI: 3D regional ventilation image that may be correlated with severity of the disease and help guiding the therapeutic care.
- Lung segmentation: more robust and faster methods to delineate the lungs and extract motion masks necessary for thoracic-image registration.
- Vascular-tree segmentation: tools to automatically detect and localize PE events in regards to the pulmonary vascular tree from CTPA
- SARCOV-IA: using thoracic CT examination from the HCL, the sub-project ambitions to propose a semi-automatic tool for the assessment of the sarcopenia and to establish a correlation between sarcopenia and the risks associated with ICU of patients.
- Radiomics: annotated database for the community, a prognostic model of COVID-19 outcome, a framework and a strong expertise for future respiratory tropic virus outbreak.
Beside COVID-19 pandemic, we think that methods developed in this group may also be useful for more conventional lung-related diseases (COPD, etc.) and generic thoracic CT image processing.
WP 2: Neuro-covid
(leaders C. Lartizien and C. Frindel)
Initial analyses of the SFNR database MRI images (based on 37 patients) reveal 8 distinctive neurological patterns of signal abnormalities including intracerebral haemorrhagic lesions (54% of patients), signal abnormalities localized in the medial temporal lobe (43% of patients), and white matter microhaemorrhages in (24% of patients) and non-confluent multifocal hyperdense lesions on FLAIR and diffusion sequences; 76% of the patients were associated with one neuroimaging pattern, 19% with two and 5% with three patterns.
Our purpose is to support the exploration of this database, as well as another database (APHP) of non-severe COVID -19 patients and in particular to carry out the necessary post-treatments to extract higher level information: structural information by extraction of nerve fiber bundles (tractography) and anatomical information by registration with reference atlases, lesion load and localization.
Three axes will be investigated according to the literature:
- Microhaemorrhages have been identified on FLAIR and diffusion MRI sequences. These microbleeds, in addition to be small, present subtle contrast so that they are currently difficult to detect and segment. Our purpose is to develop an automatic segmentation pipeline, mainly based on machine learning, to perform this task.
- Evaluate the vascular tree. Scientists suggest that COVID-19 is more likely to be a vascular disease than a respiratory one. This suggests that it could be interesting to co-localize brain lesions with regards to the cerebral vascular tree. Our purpose is thus to develop methods to extract this vascular tree from magnetic resonance angiography (MRA).
- Evaluate the olfactory nerve track. Loss of sense of smell (anosmia) and/or taste (agueusia) have been frequently observed in patients with COVID-19. These symptoms may be persistent and related to minor neurological manifestations. Anosmia could result from damages to the tissues surrounding the olfactory neurons and the olfactory bulb could be a gateway for the virus to the brain. Further investigation is required to verify this hypothesis, including the co-location of these lesions with respect to the white matter bundles of the olfactory nerve.
Expected results
- Lesion detection : unsupervised machine learning algorithms for detecting microhaemorrhages in FLAIR and SWI images
- Cerebral vascular tree segmentation: more robust and faster methods to segment the cerebral vasculature from MRA
- Tractography of the olfactive bulbe : tool to automatically extract the olfactory tracts
- Neuro-covid Pattern : preliminary analysis of the pathological patterns derived from the imaging biomarkers extracted (lesion load and localization with regard to the vascular tree and olfactory tracts).
WP 3 (transverse): IT developments
(leader F. Cervenansky)
The objective is to set up a platform that will allow a secured access to COVID-19 image databases and deployment of learning algorithms. Given that these databases contain sensitive but anonymised patient information (several types of images + patient EHR), it is essential to guarantee the legal and regulatory framework surrounding access to the data (labelled server for storage of medical data / RGPD etc.). The data cannot transit on calculation nodes outside the laboratory without the risk of breaking the traceability of the data. It is therefore necessary to use the laboratory's GPU-related computing resources.
The info-dev team of CREATIS will ensure the deployment of algorithms (via container technologies) on the calculation servers, which alone will have access to patient data. A monitoring and control infrastructure must be set up to ensure a sufficient level of service for a research use (rapid evolution of scripts, complex AI method). This essential porting and administration of these scripts will require to have interaction with researchers involved in the development of image processing, especially deep learning algorithms, in order to detect problems upstream.
Another research axis of AI that could be investigated in strong collaboration with participants of the two other work packages is that of federated learning which encompasses methods to train machine learning models on several sites respecting the personal nature of the medical data. Such developments are crucial to include new sources of distributed data (e.g. PACS or warehouses from different clinical centres) in the perspective of multicentric clinical studies.
Fundings
We recently obtained different support fundings:
- From the CARE (Comité, Analyse, Recherche, et Expertise) entity and the CNRS (as a support to the GDR ISIS initiative) to hire a one-and-a-half year engineer dedicated to the development of the platform that will allow a secured access and the deployment of learning algorithms from the databases linked to our project. Part of this funding has also been used to purchase a computing server that will be used by the engineer. The engineer will be supervised by Info-dev service of CREATIS, in particular by Frederic Cervenansky.
- From the IDEX Lyon, to hire a 6-month clinical research assistant who was in charge of collecting the pulmonary CT scans and HER of CHU St Etienne.
Complementary skills from people in different teams
Data and medical expertise will be provided by radiologists and ICU doctors from MAGICS and MYRIAD teams.
TOMORADIO team brings strong experience in ventilation imaging from CT scans, as well as in pulmonary segmentation and registration for lung-cancer treatment. Similar skills were also developed by MYRIAD team members within the framework of research on acute respiratory distress syndrome. The two subgroups now tightly collaborate within the CTVI and lung-segmentation work packages.
MYRIAD team has quickly growing skills in artificial intelligence and is involved in all tasks requiring automatic segmentation and classification for pulmonary and brain imaging applications.
Engineers from CREATIS IT team (info-dev) will play a key role in the maintenance, development (traceability, remote access, etc.) and durability of the various databases (neuro, cardiac, pulmonary): images and metadata. They will also have strong interactions for the development and deployment of algorithms workflows on computing resources (HPC and/or HTC) as well as for the relevant visualization of results. Their actions will have the will and the necessity to federate the developments of the different axes if possible.
Results
Clinical and preclinical studies
- COVID-CTPRED 2020, clinicalTrials NCT04377685
- 800 patients (419 COVID+). Complete.
- 800 patients (419 COVID+). Complete.
- CT4ARDS 2020, clinicalTrials NCT03870009
- Acute respiratory distress syndrome, 2 CT/patient.
- 31 patients (11 COVID+). Complete.
- Lungs segmented by experts in all images.
- Acute respiratory distress syndrome, 2 CT/patient.
- VT4COVID 2020, clinicalTrials NCT04349618
- 215 patients. Complete.
- 215 patients. Complete.
- CT4ARDS-2 2020, clinicalTrials NCT06113276
- Acute respiratory distress syndrome, 4 CT/patient.
- >160 patients COVID+. Recruiting.
- Lungs segmented by experts in all images.
- Acute respiratory distress syndrome, 4 CT/patient.
- PK TCM, 2021
- Images PET
- 21 pigs. Complete
- Images PET
- PK DV, 2023
- Images PET-CT.
- 11 pigs. Complete.
- Images PET-CT.
- PK HIDRA, 2023
- Images PET-CT.
- 10 pigs. Complete
- Images PET-CT.
Publications
Articles in journals
- Jean-Christophe Richard, Nicolas Terzi, Hodane Yonis, Fatima Chorfa, Florent Wallet, Claire Dupuis, Laurent Argaud, Bertrand Delannoy, Guillaume Thiery, Christian Pommier, Paul Abraham, Michel Muller, Florian Sigaud, Guillaume Rigault, Emilie Joffredo, Mehdi Mezidi, Bertrand Souweine, Loredana Baboi, Hassan Serrier, Muriel Rabilloud, Laurent Bitker, on behalf of the VT4COVID collaborators, Ultra-low tidal volume ventilation for COVID-19-related ARDS in France (VT4COVID): a multicentre, open-label, parallel-group, randomised trial, The Lancet, Respiratory Medicine, 11:11, 991-1002, November 2023, DOI: 10.1016/S2213-2600(23)00221-7
- Francois Dhelft, Sophie Lancelot, William Mouton, Didier Le Bars, Nicolas Costes, Emmanuel Roux, Maciej Orkisz, Nazim Benzerdjeb, Jean-Christophe Richard, and Laurent Bitker, "Prone position decreases acute lung inflammation measured by [11C](R)-PK11195 positron emission tomography in experimental acute respiratory distress syndrome", Journal of Applied Physiology, 2023, 134:2, 467-481, DOI: 10.1152/japplphysiol.00234.2022.
- Penarrubia L., Verstraete A., Orkisz M., Dávila Serrano E.E., Boussel L., Yonis H., Mezidi M., Dhelft F., Danjou W., Bazzani A., Sigaud F., Bayat S., Terzi N., Girard M., Bitker L., Roux E., and Richard J.-C., "Precision of CT-derived alveolar recruitment assessed by human observers and a machine learning algorithm in moderate and severe ARDS ", Intensive Care Medicine Experimental, 2023, 11, 8. DOI: 10.1186/s40635-023-00495-6.
- Jean-Christophe Richard, Florian Sigaud, Maxime Gaillet, Maciej Orkisz, Sam Bayat, Emmanuel Roux, Touria Ahaouari, Eduardo Davila, Loic Boussel, Gilbert Ferretti, Hodane Yonis, Mehdi Mezidi, William Danjou, Alwin Bazzani, Francois Dhelft, Laure Folliet, Mehdi Girard, Matteo Pozzi, Nicolas Terzi, Laurent Bitker, "Response to PEEP in COVID-19 ARDS patients with and without extracorporeal membrane oxygenation. A multicenter case-control computed tomography study", Critical Care, 26, 195, 2022, DOI: 10.1186/s13054-022-04076-z.
- Laurent Bitker, François Dhelft, Sophie Lancelot, Didier Le Bars, Nicolas Costes, Nazim Benzerdjeb, Maciej Orkisz and Jean-Christophe Richard, "Non-invasive quantification of acute macrophagic lung inflammation with [11C](R)-PK11195 using a three-tissue tissue compartment kinetic model in experimental acute respiratory distress syndrome", European Journal of Nuclear Medicine and Molecular Imaging, 2022, 49, pages 2122–2136, DOI: 10.1007/s00259-022-05713-z.
- Ludmilla Penarrubia, Nicolas Pinon, Emmanuel Roux, Eduardo Enrique Dávila Serrano, Maciej Orkisz, and David Sarrut, Improving motion-mask segmentation in thoracic CT with multi-planar U-nets, Medical Physics, 49, 420-431, 2022, DOI: 10.1002/mp.15347.
- Laurent Bitker; Nadia Cristinne Carvalho; Sacha Reidt; Christoph Schranz; Dominik Novotni; Maciej Orkisz; Eduardo Davila Serrano; Jean-Pierre Revelly; Jean-Christophe Richard, Validation of a novel system to assess end-expiratory lung volume and alveolar recruitment in an ARDS model, Intensive Care Medicine Experimental, 9, 46, 2021, DOI: 10.1186/s40635-021-00410-x.
- Louis Chauvelot, Laurent Bitker, François Dhelft, Mehdi Mezidi, Maciej Orkisz, Eduardo Davila Serrano, Ludmilla Penarrubia, Hodane Yonis, Paul Chabert, Laure Folliet, Guillaume David, Judith Provoost, Pierre Lecam, Loic Boussel, Jean-Christophe Richard, Quantitative-analysis of computed tomography in COVID-19 and non COVID-19 ARDS patients: a case-control study, Journal of Critical Care, 60:169-176, 2020, DOI : 10.1016/j.jcrc.2020.08.006.
Communications at conferences - articles
- M. Shekarnabi, T. Ahaouari, J.C. Richard, S. Bayat, M. Orkisz, “CT registration-derived biomarkers of recruitability in ARDS”, IEEE International Symposium on Biomedical Imaging, Cartagena de Indias, Colombia, April 18-21 2023, pp. 1-5, DOI: 10.1109/ISBI53787.2023.10230395.
- Eduardo E. Dávila Serrano, François Dhelft, Laurent Bitker, Jean-Christophe Richard, and Maciej Orkisz. “Software for CT-image Analysis to Assist the Choice of Mechanical-ventilation Settings in Acute Respiratory Distress Syndrome”, International Conference on Computer Vision and Graphics, Warsaw, Poland, September 14-16, 2020, Leszek J Chmielewski, Ryszard Kozera, Arkadiusz Orłowski Eds., Springer, LNCS 12334, pp 48-58, DOI: 10.1007/978-3-030-59006-2_5.
Communications at conferences - abstracts
- Mehdi Shekarnabi, Florian Sigaud, Maxime Gaillet, Alicia Guillien, Emmanuel Roux, Touria Ahaouari, Edouardo Davila, Loic Boussel, Gilbert Ferretti, Hodane Yonis, Mehdi Mezidi, William Danjou, Alwin Bazzani, François Dhelft, Laure Folliet, Mehdi Girard, Valérie Siroux, Laurent Bitker, Nicolas Terzi, Maciej Orkisz, Jean-Christophe Richard, Sam Bayat, “Phenotypes of functional CT imagining predict clinical outcome in ARDS patients”, European Respiratory Society (ERS) International Congress, Milan, Italy, Sept. 2023, European Respiratory Journal, Vol 62 Issue suppl. 67 OA4955, 2023 DOI: 10.1183/13993003.congress-2023.OA4955.
- JC Richard, H Yonis, F Sigaud, M Mezidi, E Davila, M Orkisz, E Roux, M Gaillet, S Bayat, N Terzi, L Bitker, “Response to PEEP assessed on computed tomography in COVID-19 ECMO patients”, Réanimation 2022 Paris 22-24/06/2022.
- Pariente T., Osowiechi D., Delorme J., Granier L., Kandji M., Holdner A., Peyrat L., Grenier T., Falandry C., Pialat J.-B., “Dévelopement d’un reseau de neurons et évaluation de l’intérêt pronostique des marqueurs TDM de sarcopénie”, JFR 2022.