Abstract
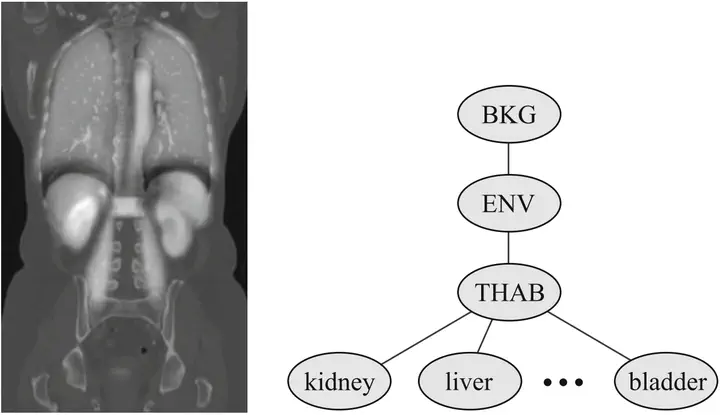
We propose a generic method for the automatic multiple-organ segmentation of 3D images based on a multilabel graph cut optimization approach which uses location likelihood of organs and prior information of spatial relationships between them. The latter is derived from shortest-path constraints defined on the adjacency graph of structures and the former is defined by probabilistic atlases learned from a training dataset. Organ atlases are mapped to the image by a fast (2+1)D hierarchical registration method based on SURF keypoints. Registered atlases are also used to derive organ intensity likelihoods. Prior and likelihood models are then introduced in a joint centroidal Voronoi image clustering and graph cut multiobject segmentation framework. Qualitative and quantitative evaluation has been performed on contrast-enhanced CT and MR images from the VISCERAL dataset.