Abstract
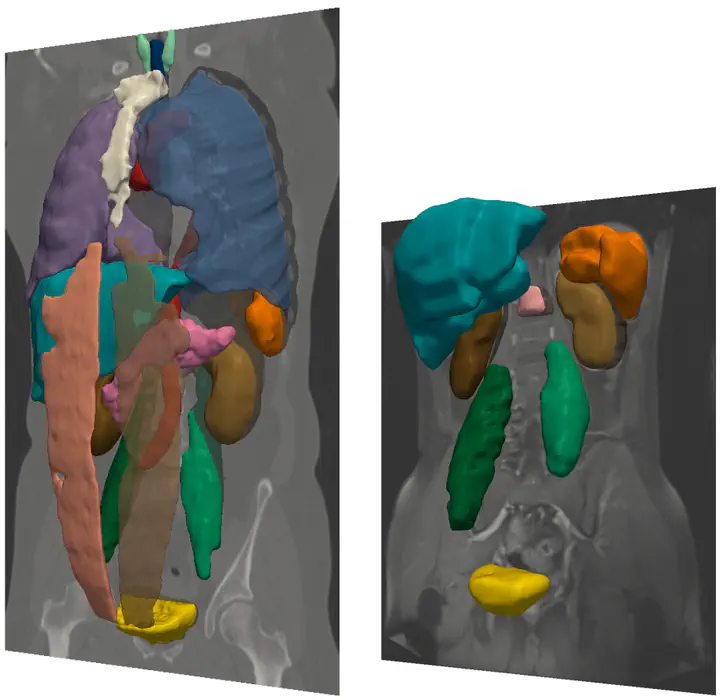
We propose an automatic multiorgan segmentation method for 3D radiological images of different anatomical content and modality. The approach is based on a simultaneous multilabel Graph Cut optimization of location, appearance and spatial configuration criteria of target structures. Organ location is defined by target-specific probabilistic atlases (PA) constructed from a training dataset using a fast (2+1)D SURF-based multiscale registration method involving a simple 4-parameter transformation. PAs are also used to derive target-specific organ appearance models represented as intensity histograms. The spatial configuration prior is derived from shortest-path constraints defined on the adjacency graph of structures. Thorough evaluations on Visceral project benchmarks and training dataset, as well as comparisons with the state of the art confirm that our approach is comparable to and often outperforms similar approaches in multiorgan segmentation, thus proving that the combination of multiple suboptimal but complementary information sources can yield very good performance.