Purpose and context
The objective is to extract quantitative parameters from the 3D images of the trabecular bone network. With 2D images such as used in histomorphometry, stereology methods allowing to infer 3D parameters from 2D measurements, require a priori models of the investigated structure. Bone micro-architecture is typically assumed to follow a parallel-plate model, which is not necessarily true. With 3D images, such an assumption is no more necessary and model-independent methods may be used. In addition, 3D images allow extracting information about the morphology, the anisotropy and the topology of the trabecular network.
Methods
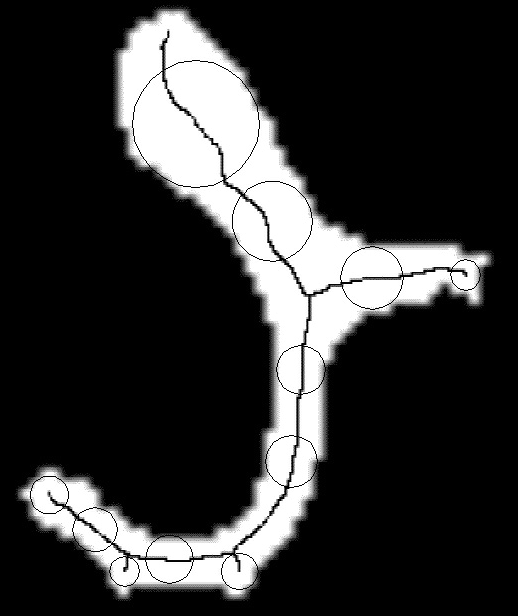
In addition to standard quantification based on the MIL (Mean Intercept Length) method, several quantification methods were developed. Direct thickness measurement: The measurement of the direct trabecular thickness, proposed by Hildebrand and Ruegsegger in a continuous framework was implemented using 3D discrete distance maps. The method provides a 3D map of the local trabecular thickness. Local geometric analysis : We proposed an original method to evaluate locally the geometry of complex structures. The method is based on 3D medial axis representation followed by topologic analysis (collaboration D. Attali, LIS, Grenoble)
Results
Direct thickness:
|
(a) |
(b) |
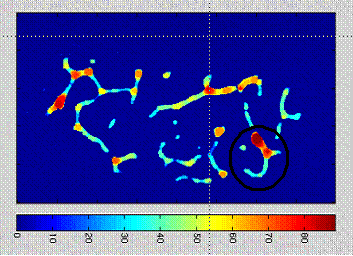
a) Illustration of the local trabecular thickness on a 2D slice extracted from the 3D image : the trabecular thickness at one point is defined as the diameter of the maximal ball including that point. b) Color-coded representation of a slice of the 3D trabecular thicknessl maps
Geometric parameters :
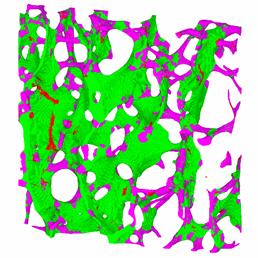
The method allows to classify each voxel of the 3D image according to its geometry. The method was first tested on simulated phantoms made of plates and cylinders, and then applied to experimental data. Despite of the complexity of the trabecular bone network, the method provided good results (see figure below). As compared to previous approaches, the method allows to classify the entire volume and only its skeleton, and it appears to be robust to small irregularities on the bone surface.
|
(a) |
(b) |
Illustration of the local classification of each voxel of a trabecular bone 3D image : a) original image, b) Color-coded representation of the classified image : green : plate , pink : rods, red : nodes
The method was applied to trabecular bone images in human