- Participants
P. Clarysse, N. Duchateau (eq. 1), G. K. Rumindo (PhD student)
P. Croisille, M. Viallon (eq. 5)
- National and international collaborations
J. Ohayon (TIMC-IMAG, Grenoble, France)
P. Bovendeerd (TU/e, Eindhoven, The Netherlands)
V. Wang, M. Nash, A. Young (Auckland Bioengineering Institute, New Zeland)
- Question
How to accurately characterize, track and predict the evolution of functional cardiac recovery following coronary revascularization ?
- Action objective
Our approach is based on the personalization of a 3D finite element (3D-FE) biomechanical model of the beating heart from dynamic imaging data. Cardiac functional status is evaluated through mechanical properties provided by the model. Properties are statistically analyzed on population of 3D-FE models, identified from clinical data or artificially produced, to determine those that better characterize the pathology. Therefore, it consists in the three stages:
- To personalize a 3D-FE model of the beating heart from dynamic cardiac imaging data,
- To produce model populations with different characteristics in terms of mechanical properties and pathology,
- To determine the parameters that best characterize the regional myocardial function and its evolution during the course of the pathology and following revascularization.
- Method
- Personalization through an inverse bio-mechanical approach. The 3D-FE model is based on the isotropic transverse quasi-uncompressible material law of Fung adapted to the myocardium by Guccione et al. for the passive part and on a stretch dependent active force law. FE implementation is performed in Abaqus software. Personalization is achieved through non-linear optimization under pressure-volume relation and/or imaging based deformation constraints,
- Generation of model populations. From a geometrical model basis, new cases are produced through randomization within the range of variation of the mechanical and infarct parameters,
- Identification of function markers. Statistical integrative data analysis is performed on virtual and clinical cohorts to determine the most discriminant parameters for the assessment of the passive and active myocardial contractile ability (see action ‘Parameter analysis and marker extraction’.
- Results and illustrations
Figure 1 shows some examples of 3D-FE left ventricular (LV) models with lesions of various shape, size, location and mechanical parameters from our synthetic model database [RUMI-17], [RUMI-17a].
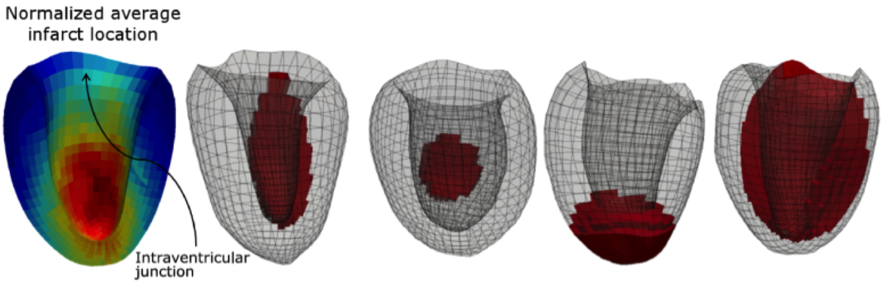
Figure 1. Average distribution of the lesions in a database of 200 synthetic pathological LVs (left, the most represented pathological locations are in red) and four examples of lesions with different shape, size and location.
Figure 2 shows the cartographies of 8 classical strain parameters produced by the 3D-FE model where the pathology has a given geometrical and mechanical configuration. Principal and radial strains, and some strain invariants correctly delineate the lesion [RUMI-17a]. Fiber strain (i.e. strain along fiber) has been particularly studied by mixing functional data from MRI tagging and structural data from Diffusion tensor MRI in partnership with the cardiac research group at the Auckland Bioengineering Institute [WANG-16].
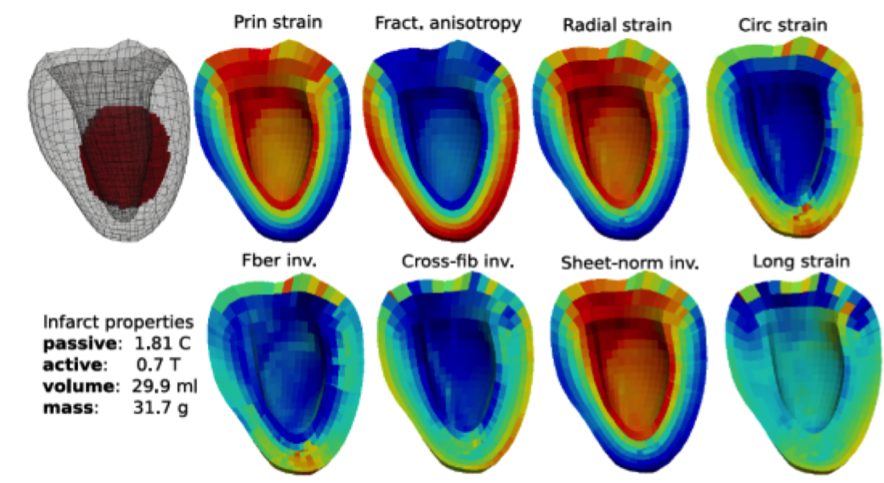
Figure 2. 3D cartography for 8 strain parameters produced by a 3D-FE LV model where the injured zone has stiffness 1.8 higher than the normal value and activation reduced to 70%.
- Financial supports
- Projet Marie-Curie VPH-CaSE (VPH-Cardiovascular Simulation and Experimentation for Personalised Medical Devices, H2020-MSCA-ITN-2014) : financement thèse G. K. Rumindo.
- Projet PALSE-IMPULSION CardioMIX (Mixing and learning multiple heterogeneous descriptors of cardiac function : integrating clinical constraints and expectations), N. Duchateau.
- Projet ANR MOSIFAH (Modélisation et simulation multimodales et multiéchelles de l'architecture des fibres myocardiques du cœur humain, ANR-13-MONU-0009), Y. Zhu.
- Bibliographic references
- Journals
[WANG-16] V. Wang, C. Casta, Y. M. Zhu, B. Cowan, P. Croisille, A. Young, P. Clarysse, and M. Nash, "Image-based investigation of human in vivo myofibre strain," IEEE Transactions on Medical Imaging, vol. PP, no. 99, pp. 1-1, 2016.
- Conferences
[RUMI-16] G. K. Rumindo, J. Ohayon, M. Viallon, M. Stuber, P. Croisille, and P. Clarysse, "Comparison of different strain-based parameters to identify human left ventricular myocardial infarct: a three-dimensional finite element study," Computer Methods in Biomechanics and Biomedical Engineering (CMBBE), Tel-Aviv, Israel, 2018, pp. 161-169.
[RUMI-17] G. K. Rumindo, N. Duchateau, P. Croisille, J. Ohayon, and P. Clarysse, "Strain-Based Parameters for Infarct Localization: Evaluation via a Learning Algorithm on a Synthetic Database of Pathological Hearts," in Functional Imaging and Modeling of the Heart (FIMH), Toronto, Canada, 2017, pp. 106-114.
[RUMI-17a] G. K. Rumindo, N. Duchateau, M. Viallon, P. Croisille, J. Ohayon, and P. Clarysse, "Generation of a synthetic database of heart models to evaluate strain-based infarct detection algorithms," in Recherche en Imagerie et Technologies pour la Santé, Lyon, France, 2017.