Context
Cerebral emboli are gaseous or solid particles circulating in the cerebral blood flow. They are among the primary causes of stroke, a leading cause of death and disability worldwide [1]. Portable Transcranial Doppler (TCD) is the only modality that enables real-time, long-term monitoring of emboli in the cerebral arteries. Some examples of signals spectrograms are given Figure 1. Recent studies have shown that deep learning techniques are effective for classifying embolic signals [2].
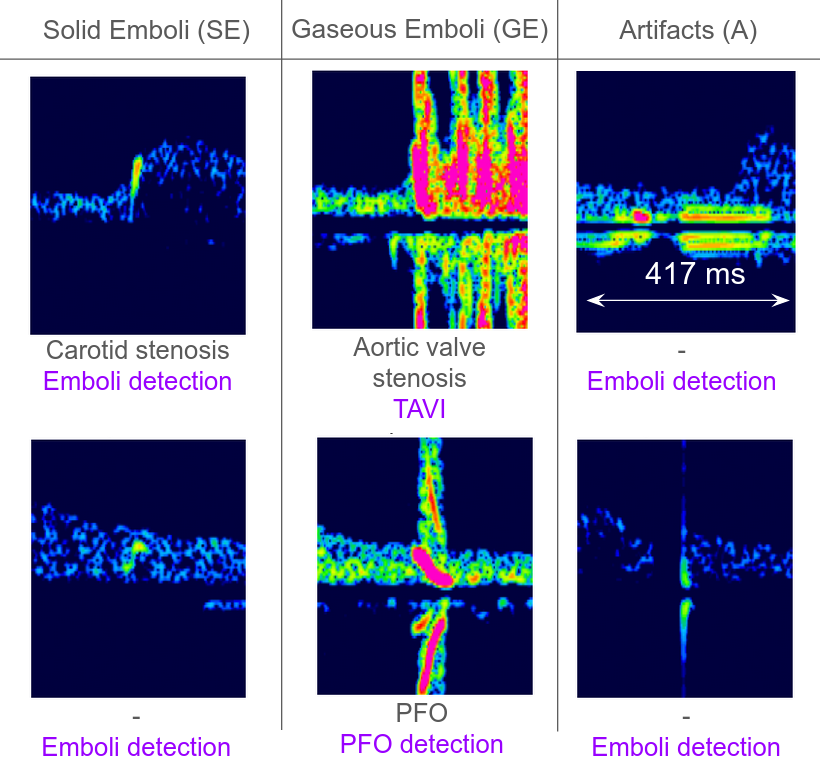
Figure 1. Spectrograms of TCD signals from different classes. For each image, the linked pathology and the reason for the TCD examination are indicated when available.
However, current deep learning models are typically trained on fixed, population-based datasets whereas emboli are encountered in diverse clinical contexts and patient populations, as they are emitted in various scenarios such as different pathologies or during cardiac surgeries.
On the other hand, continual learning (CL) is a recent field of machine learning research focused on developing methods that enable models to continuously learn and adapt to new data without forgetting previously acquired knowledge [3] (cf. Figure 2.). These methods could enable the deployment of a pre-train model from a fixed database that will then specialize locally in each service while keeping a global overview of the different embolic signals.
Away from the emboli classification example, this problem is general to all AI-based models deployed in clinics and is a promising research direction in the medical domain [4].
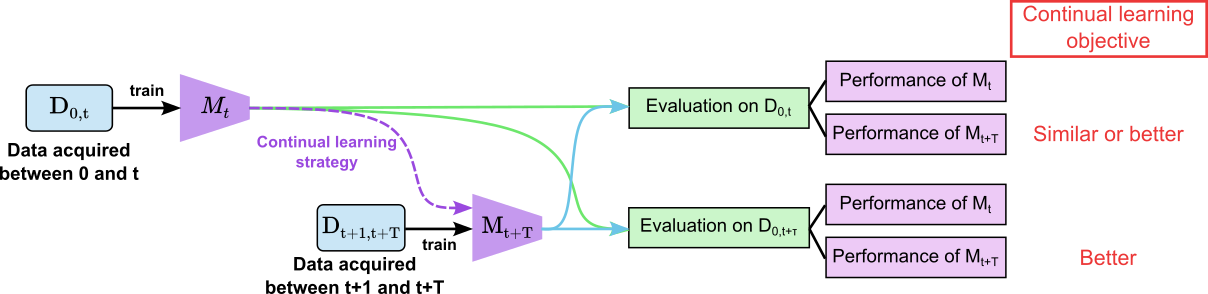
Figure 2. General principle of continual learning, where M is the trained model and D the dataset
Objectives
- Study the literature about continual learning in light of the medical context (data privacy) and the objective at aim (specialization by service, a new data is a new patient)
- Adapt a method to the context provided above, with one of the following possible directions, depending on your interest and on the bibliography:
- Replay-based CL approaches taking advantage of uncertainty quantification for memory creation and updates ;
- Using unannotated data, i.e. unsupervised continual learning ;
- Exploring the impact of noisy-annotations on supervised CL.
- Evaluate the method on the private TCD dataset and on a publicly available dataset like PTB (electrocardiograms).
Expected deliverables
- Methodological contribution on one of the directions mentioned above
- Documented code in Python using Pytorch
- Potentially writing a conference article depending on progress
Profile
- Student in 2nd year of Master (or equivalent) with skills in applied mathematics and/or informatics.
- Good machine and deep learning background.
- Be familiar with Python programming.
- Proficiency in English is essential (be able to read bibliography, write reports, work in an international environment, attend and give presentations in English).
- Be motivated to work on medical applications and on an exploratory question.
Supervision
Mathilde Dupouy (PhD student)
Philippe Delachartre (Professor)
Blaise Kévin Guépié (Associate professor)
Yamil Vindas Yassine (Postdoctoral researcher)
Practical information
Site : CREATIS lab, campus de la DOUA, Villeurbanne (Lyon)
Gratification : ~650 €/month
Duration : ~ 6 months, starting Feb./March (3 weeks of closure in August)
Language : English or French
Contact : send CV and motivation letter to mathilde.dupouy@creatis.insa-lyon.fr
References
[1] J. D. Steinmetz et al., “Global, regional, and national burden of disorders affecting the nervous system, 1990–2021: a systematic analysis for the Global Burden of Disease Study 2021,” Lancet Neurol., vol. 23, no. 4, pp. 344–381, Apr. 2024, doi: 10.1016/S1474-4422(24)00038-3.
[2] Y. Vindas, E. Roux, B. K. Guépié, M. Almar, and P. Delachartre, “Guided deep embedded clustering regularization for multifeature medical signal classification,” Pattern Recognit., vol. 143, p. 109812, Nov. 2023, doi: 10.1016/j.patcog.2023.109812.
[3] S. Farquhar and Y. Gal, “Towards Robust Evaluations of Continual Learning,” Jun. 26, 2019, arXiv: arXiv:1805.09733. doi: 10.48550/arXiv.1805.09733.
[4] D. Kiyasseh, T. Zhu, and D. Clifton, “A clinical deep learning framework for continually learning from cardiac signals across diseases, time, modalities, and institutions,” Nat. Commun., vol. 12, no. 1, p. 4221, Jul. 2021, doi: 10.1038/s41467-021-24483-0.
