- Participants
Nicolas Duchateau, Maxime Di Folco, Benoit Freiche, Patrick Clarysse.
- National and international collaborations
Universitat Pompeu Fabra, Barcelone, ES (Sergio Sanchez, Gemma Piella, Bart Bijnens)
CHU Nice, FR (Pamela Moceri)
Question
How to integrate a large amount of data, often of heterogeneous and complex types ? Which impact for the understanding of diseases ?
- Objective
Further developing machine learning methods on heterogeneous and high dimensional data, and using them to (i) better represent the distribution of a population and (ii) evaluate the relative importance of the evaluated features and point out new interactions between such features.
- Method
The methodology builds upon unsupervised manifold learning that also reduce the dimensionality of data. The data variability is then studied in the low-dimensional space or after reconstruction in the input space, depending on the application needs.
We have contributed to the development of an unsupervised version of the Multiple Kernel Learning algorithm (originally introduced in [Lin:2011]). An affinity matrix is built for each feature via a appropriate kernel, and the algorithm jointly optimizes the relative weight of these features and a transformation matrix to a space of lower dimensionality. We also investigate strategies that better take into account the links between the inputs descriptors, such as their correlation and potential redundancy [DiFolco:2019].
- Results
We tested the multiple kernel methodology on different descriptors of myocardial velocity from tissue Doppler acquisitions from a stress protocol, to better characterize the heart failure with preserved ejection fraction (HFPEF) syndrome [Sanchez:2017, Sanchez:2018], and on a set of clinical and imaging decriptors from a 2D echocardiography protocol to identify differences in response to cardiac resynchronization therapy [Cikes:2019].
The links between descriptors are currently explored for cardiac shape and deformation, to study pressure and volume overload of the right ventricule from 3D echocardiographic data.
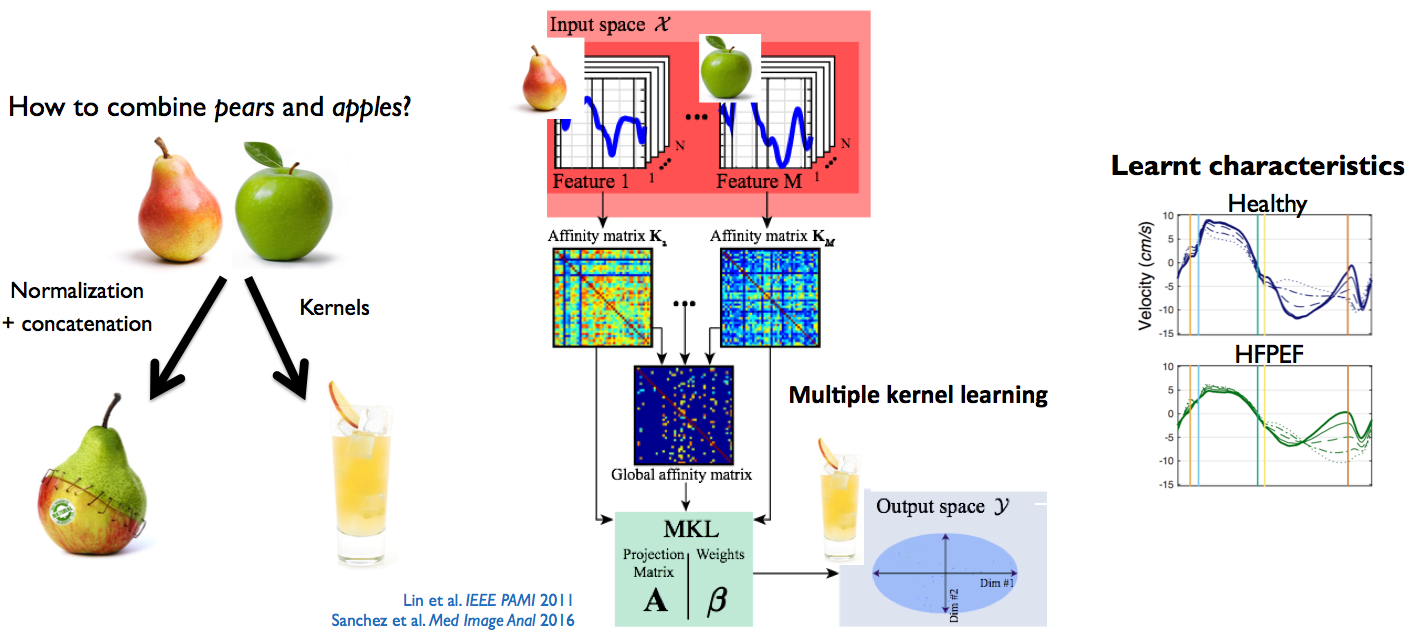
Figure 1: methodology used to better represent the data from several heterogeneous descriptors. Illustration on myocardial velocities from normal subjects and patients with heart failure and preserved ejection fraction, before and during a stress protocol. Adapted from [Sanchez:2017].
- Grants
PALSE-IMPULSION-2017 (Programme d’Avenir Lyon Saint Etienne).
VP2HF (FP7-ICT-2013-611823).
ANR JCJC MIC-MAC (ANR-19-CE45-0005).
- Publications
[Lin:2011] Lin YY, Liu TL, Fuh CS. Multiple kernel learning for dimensionality reduction. IEEE Trans Pattern Anal Mach Intell. 2011;33:1147-60.
[Sanchez:2017] Sanchez-Martinez S, Duchateau N, Erdei T, Fraser A, Bijnens B, Piella G. Characterization of myocardial motion patterns by unsupervised multiple kernel learning. Med Image Anal. 2017;35:70-82.
[Sanchez:2018] Sanchez-Martinez S, Duchateau N, Erdei T, Kunszt G, Aakhus S, Degiovanni A, Marino P, Carluccio E, Piella G, Fraser AG, Bijnens BH. Machine learning analysis of left ventricular function to characterize heart failure with preserved ejection fraction. Circ Cardiovasc Imaging. 2018;11:e007138.
[Cikes:2019] Cikes M, Sanchez-Martinez S, Claggett B, Duchateau N, Piella G, Butakoff C, Pouleur AC, Knappe D, Biering-Sørensen T, Kutyifa V, Moss A, Stein K, Solomon SD, Bijnens B. Machine-learning based phenogrouping in heart failure to identify responders to resynchronization therapy. Eur J Heart Fail. 2019;21:74-85.
[DiFolco:2019] Di Folco M, Clarysse P, Moceri P, Duchateau N Learning interactions between cardiac shape and deformation: application to pulmonary hypertension. Proc. STACOM-MICCAI, LNCS. 2019. In press.