Machine learning for medical imaging
Outlier detection model and deep unsupervised representation learning
Standard supervised learning framework is barely adapted to the detection task of subtle anomalies with varying size and shape and localisation in the organ of interest. Such supervised models indeed require to collect massive annotated image database, where the annotation task is all the more difficult and time consuming that lesions are not or barely visually detected by clinical experts in images.
Over the last research period, we proposed to tackle this problem as an outlier detection problem, which allows bypassing the need for large annotated databases and the class imbalance inherent to this detection task. Our first model combined manual engineered features to one-class SVM [Azam-2016- PlosOne].
We then presented a method for converting SVDD scores (equivalent to one-class SVM) into well-calibrated probabilities so as enable efficient post-processing and inter-individual comparison of the detection maps [El Azami, Neurocomputing 2017].
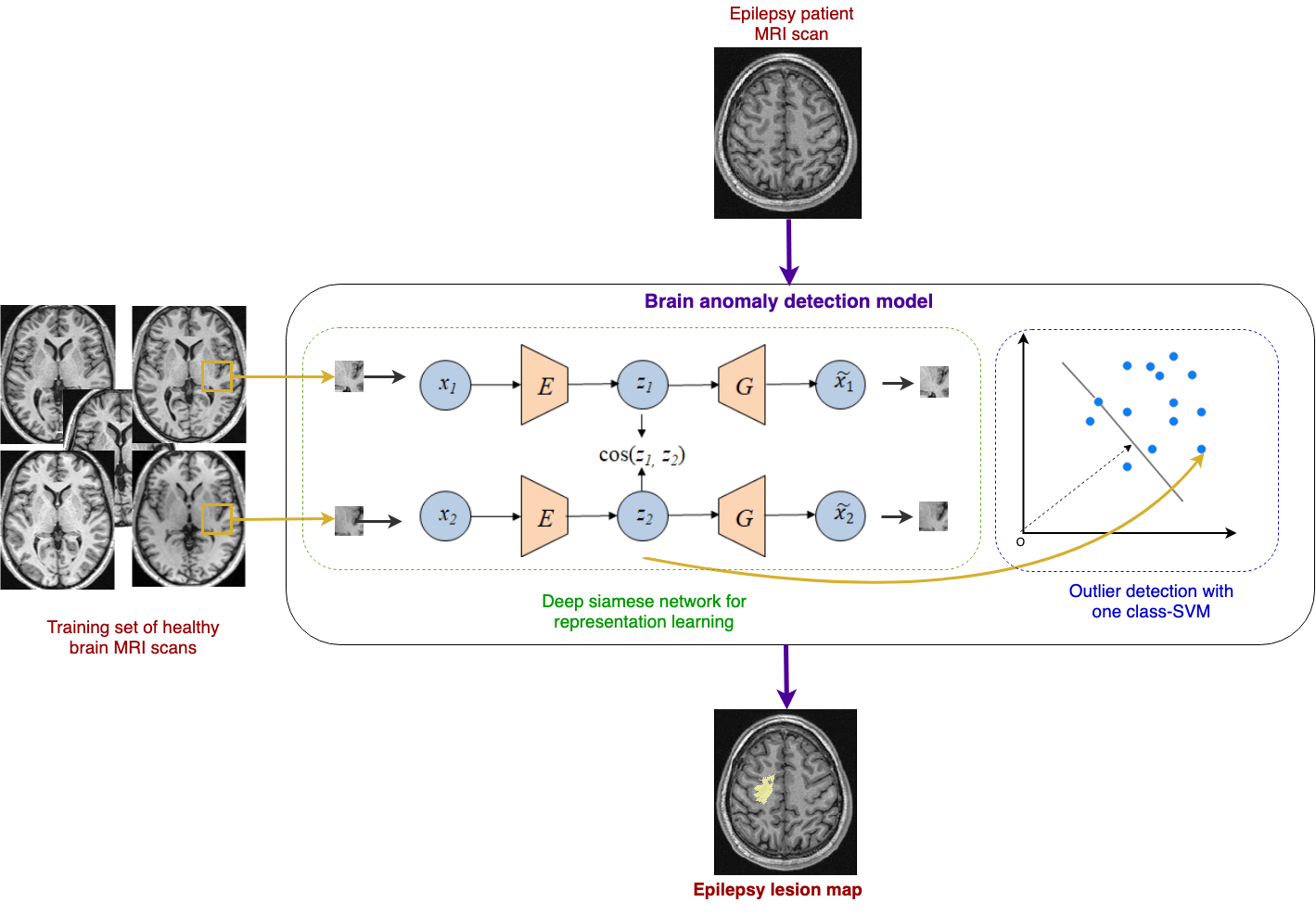
In a recent study, we proposed a novel deep unsupervised representation model based on Siamese autoencoders as an alternative to the manual feature extraction step [Alaverdyan, MEDIA 2020], [Alaverdyan, MIDL 2018], [Alaverdyan, MICCAI DLMIA 2018].
This deep representation is then coupled to a one-class SVM.
We plan to dig into this research axis by developing unsupervised representation models that better capture the inherent complex patterns of medical imaging data (eg encoding constraints loss terms in deep architecture based on anatomical or spatial localisation priors ..) as well as proposing efficient ways to encode end fuse the information provided by different imaging modalities.
Domain Adaptation
Our main methodological focus over the past 5 years was to address the issue of transfer learning, and more specifically of domain adaptation, occurring when learning is performed based on data samples following different probability distributions, as encountered with multicentric data. Novel methodological contributions in optimal transport were efficiently transferred to clinic [Gautheron-2017- GRETSI] [Gautheron-2018- ECML PKDD], and original theoretical contributions in metric learning were recently proposed [Dhouib-2018- CAp] [Dhouib-2018- NeurIPS].
Kernel machines
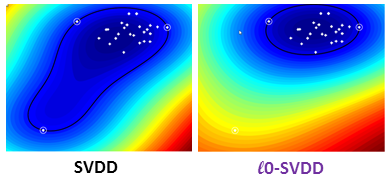 The objective of this research axis is to develop or adapt statistical learning algorithms, such as support vector machines (SVM), to the specificity of clinical imaging data which include, for instance, uncertainty about the reference training data, low sample and high dimensional data.
The objective of this research axis is to develop or adapt statistical learning algorithms, such as support vector machines (SVM), to the specificity of clinical imaging data which include, for instance, uncertainty about the reference training data, low sample and high dimensional data.
These projects are conducted in collaboration with experts in machine learning or applied mathematics.
Our contributions in this field include:
- Kernel-based learning from data with uncertain class labels [Niaf, IEEE TIP 2014]
- Robust outlier detection with SVDD in the presence of uncertain data [El Azami, ESANN 2014]
- Converting scores outputed by kernel-based outlier detection algorithm into probability estimates [El Azami, Neurocomputing 2017]