How to participate
-
Segmentation outputs
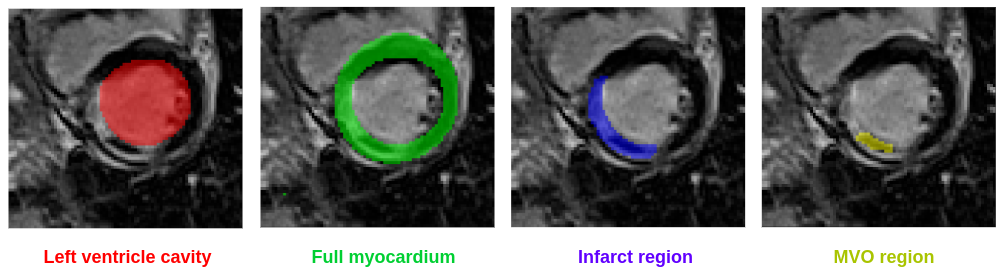
MYOSAIQ is a segmentation contest: participants are invited to develop automatic tools to segment four different cardiac structures from LGE images, i.e.
- the left ventricle cavity
- the full myocardium composed of healthy myocardial tissues with potential infarct tissues and MVO
- the infarct region composed of infarct tissues with potential MVO
- the microvascular obstruction (MVO) region (hypo-enhanced zone)
Please note that for reasons of compactness, the ground truth volumes have labels with the following intensity values:- Left ventricle cavity: pixel intensity of 1
- Healthy myocardial tissues: pixel intensity of 2
- Infarct tissues: pixel intensity of 3
- MVO region: pixel intensity of 4
The following relationships must therefore be taken into account: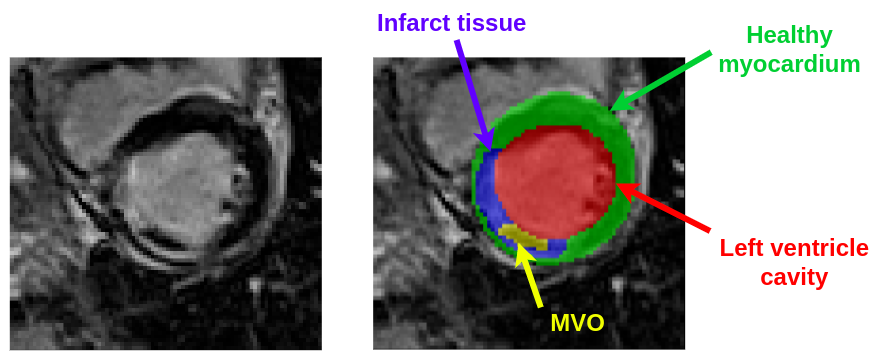
- the left ventricle cavity corresponds to the set of pixels with a value of 1
- the full myocardium corresponds to the set of pixels with values of 2, 3 or 4
- the infarct region corresponds to the set of pixels with values of 3 or 4
- the MVO region corresponds to the set of pixels with a value of 4
Clinical outputs
Each participant is also asked to calculate the volumes and associated confidence for the 4 segmented cardiac structures, namely the LV cavity, the full myocardial region, the infarct region and the MVO region. The confidence will be evaluated from the Continuous Ranked Probability Score (CRPS), inspired from the metric proposed during the Kaggle 2nd annual data science bowl. To this end, for each LGE MR image, participants must predict a cumulative probability distribution for each of the four structures mentioned above. The CRPS is computed for each structure as follows:
where P is the predicted cumulative distribution, N is the number of MR-LGE exams, V is the actual volume (in mL) and H(x) is the Heaviside step function (H(x) = 1 for x >= 0 and zero otherwise). For a better understanding, we provide in the figure below an illustration of the error term (green area) between the predicted cumulative distribution and the Heaviside step function centred on the real volume.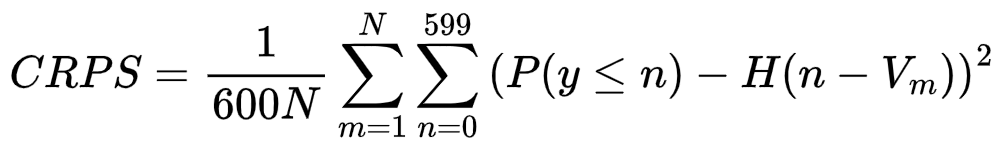
As an example, if a participant is able to estimate a volume with its confidence value for a given structure, it is straightfoward to compute the associated cumulative distribution under the Gaussian assumption where the volume and confidence values can be taken as the mean and standard deviation of the distribution. However, participants may provide distributions which do not follow a Gaussian distribution.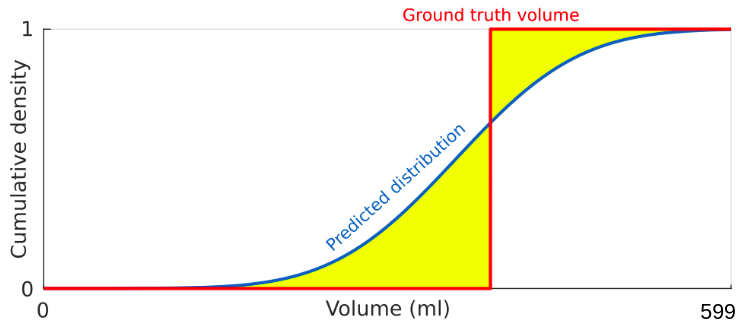
Participants who are not able to estimate the confidence in the estimate of the different volumes can still take part in the challenge. In this case, the cumulative distributions associated with each estimated volume are automatically calculated and correspond to Heaviside step functions centred on the volumes automatically computed from the segmentation estimates.Challenge rules
The training and testing dataset are composed of subfolds with images at three different phases (4-8 days, 1 month and 12 months post-MI & reperfusion) of the longitudinal evolution of the myocardial infarction.
A single method must be developed to segment the cardiac structures at the three different phases. Indeed, through the challenge we wish to validate the hypothesis that the method that will perform best globally is also the method that will perform best on the 3 phases individually.
However, as the date of the infarction is known in clinical routine, participants are allowed to use the time point information (D8, M1 or M12) to condition their method if they wish. Indeed, this information can be relevant, in particular for the segmentation of the MVO structure which is only visible in the images during a short period (a few days) following the infarction. Time information can be easily extracted by the participants from the name of the image files (e.g. the first patient of the database with an image acquired 4-8 days post-MI is named 001_D8.nii.gz).