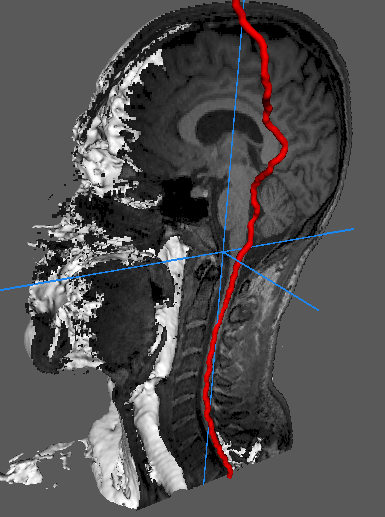
This page presents our work on spinal cord centerline localization.
Our main result is a very robust and efficient method to localize the spinal cord on 3D images.
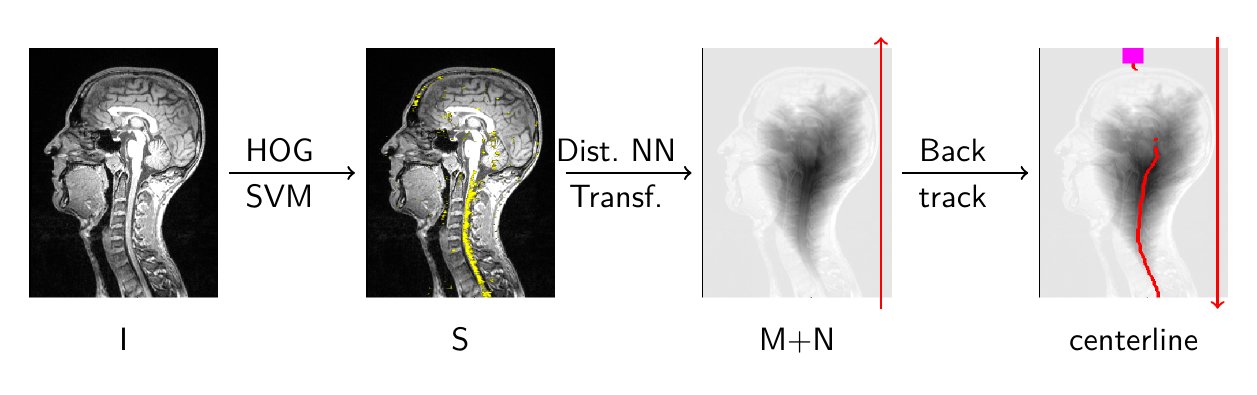
A localisation map is first computed using a HOG+SVM spinal cord detector. The centerline curve is then found as the solution of a nonlinear optimization problem. An original algorithm is proposed to find the global solution of this problem in a efficient and non iterative manner.
The method has been presented at RITS 2017
Details of the method can be found here.
Software
The binaries are availaible here for linux-x86_64, macos 10.7, macos 10.11 and windows.
It is also included and packaged in the Spinal Cord Toolbox as sct_detect_spinalcord along with a lot of spinal cord dedicated tools. It is now used for example to initialize the spinal cord segmentation tool.
Please reference the MEDIA journal article
Author
M. Sdika
Collaborations
V. Callot at CRMBM
The Neuropoly team at the Polytechnique Montréal.