Cardiac Structures Segmentation
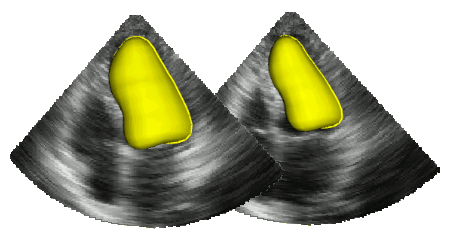
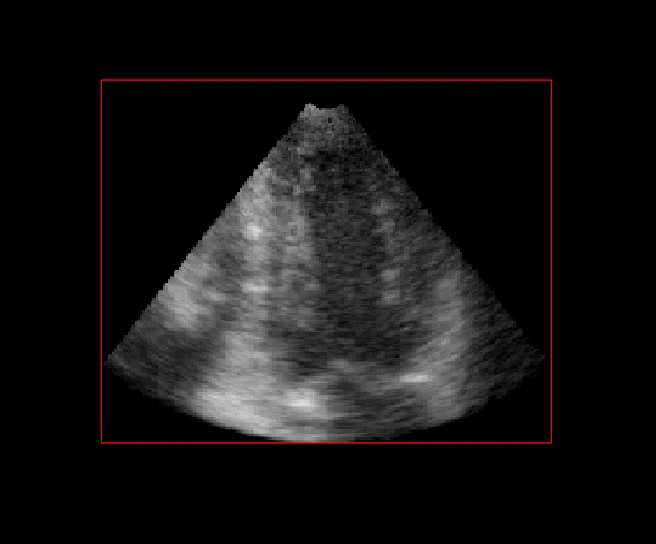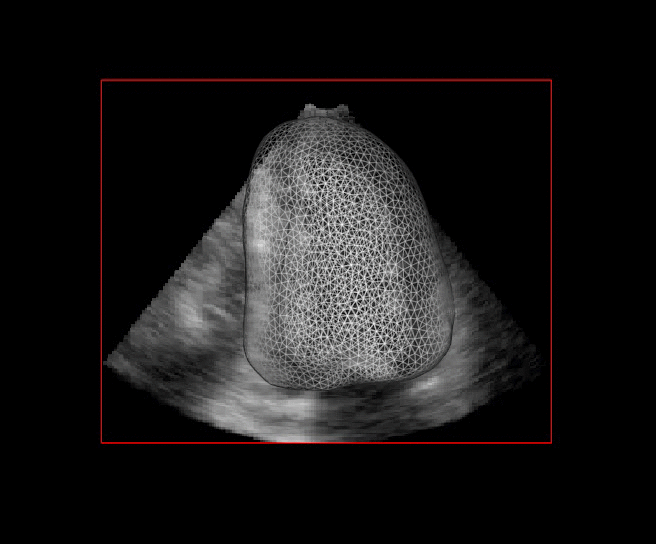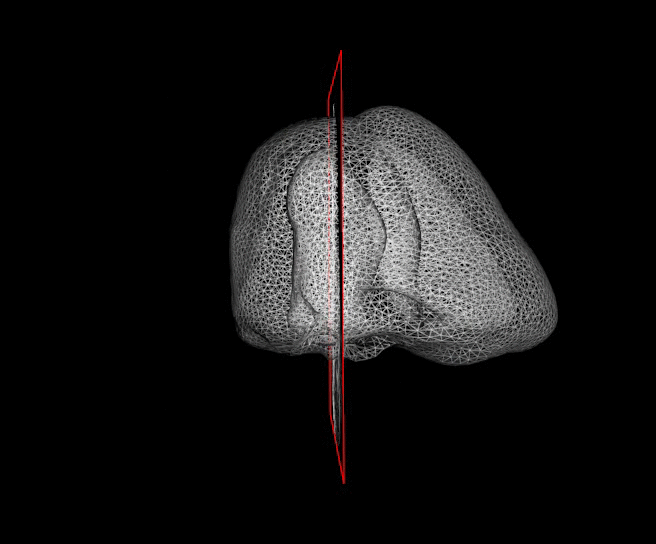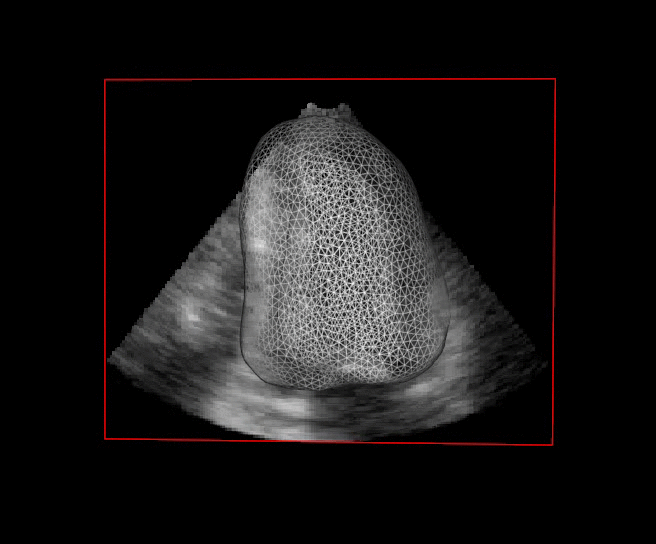
The segmentation of cardiac structures in medical imaging is a 30 years old problem that needs to be solved ! For more than 10 years, we developped innovative methods which allows a constant improvement of the overall accuracy of our segmentation performance. In particular our Shape-Based B-Spline Exlicit Active Surfaces currently outperforms other state-of-the-art methods for the automatic segmentation of left ventricle in 3D echocardiography [IEEE TMI, 2017].
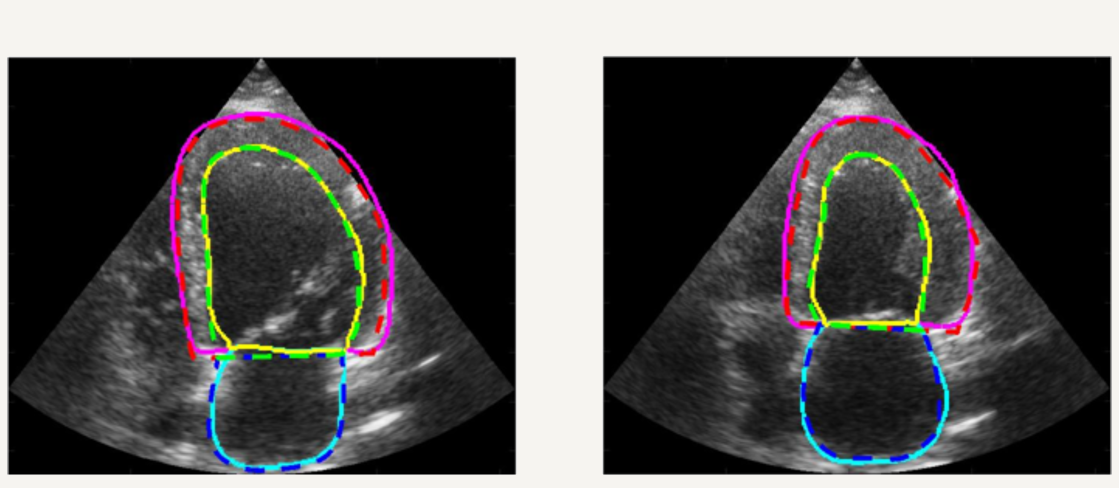
In addition, we setup a complete study for 2D echocardiography image segmentation based on the first open-access large-scale dataset composed by more than 500 patients ! Thanks to this unique dataset, we showed that an optimized U-Net method (encoder-decoder deep learning architecture) currently outperformes all the tested state-of-the-art methods and reaches accuracy below the inter-observer scores but still slight worse than the intra-observer' ones. A dedicated Girder on-line platform has been specifically setup for new result submissions where the dataset is also available for download. [IEEE TMI, 2019].
Realistic Synthetic Cardiac Sequences
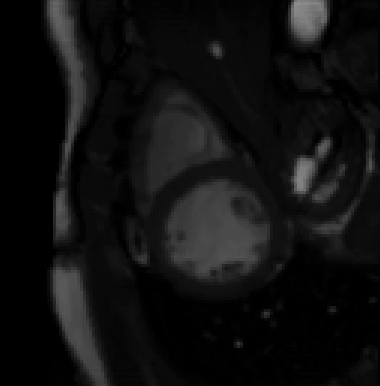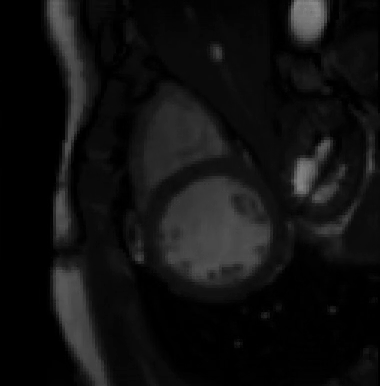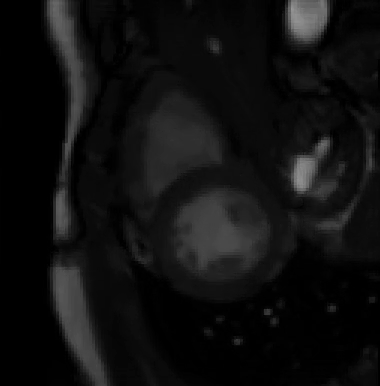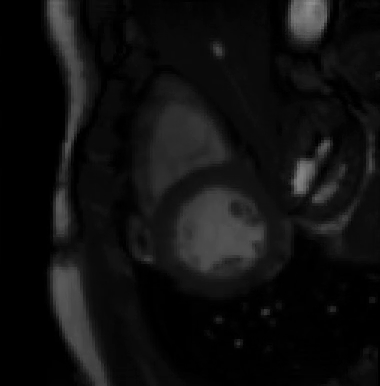
For more than four years, we are working on improving the realistic aspect of synthetic simulation of cardiac sequences for both MRI and ultrasound sequences. Such sequences are needed for an accurate evaluation of the performance of state-of-the-art methods for the myocardium strain estimation [IEEE TMI, 2017].
 |
 |
 |
Ultrafast Echocardiography
We introduced a Fourier-based technique for the reconstruction of 90o-wide echocardiography at >250 frames/s using diverging beams. Our method allows integrating motion compensation in the coherent compounding process to cope with phase delays due to myocardial motion [IEEE TUFFC, 2016].
Open science initiatives
The reproducibility of results and the ability to compare the performance of methods in a fair manner have become an absolute necessity in medical image analysis because of the intensive publication of articles in this domain. For more than 3 years, we are actively organizing and participating to open science projects to provide the community with the essential tools, platforms and databases so to continue to make major advances in our domain.
 |
CETUS: Challenge on Endocardium Three-dimensional Ultrasound Segmentation (MICCAI 2014) - This projects led to an open database of 45 3D cardiac ultrasound sequences with manual annotations. An online evaluation platform remains open for new submissions - Web link - [IEEE TMI, 2016] |
 |
PICMUS: Plane-wave Imaging Challenge in Medical ultraSound (IEEE IUS 2016) - This projects led to an open database consisting of synthetic, in vitro and in vivo data acquired using plane-waves with different steering angles - Web link |
 |
ACDC: Automated Cardiac Diagnosis Challenge (MICCAI 2017) - This projects led to an open database of 150 multi-slices cardiac MRI sequences with manual annotations. An online evaluation platform remains open for new submissions - Web link |
 |
STRAUS: STrain Assessment in UltraSound - This projects aims at providing a simulation framework for the generation of realistic 3D synthetic cardiac ultrasound and magnetic resonance (both cine and tagged) image sequences. A dataset of 18 virtual is publicly available for benchmark - Web link - [IEEE TMI, 2017] |