inTag : Cardiac MRI tagging analysis toolbox - frequently asked questions
inTag V1.0 plugin: what are the differences between the demo and the registered versions?
24/12/09
inTag demo:
-all features are enabled for testing purposes
-can only process the provided datasets
-«inTag DEMO» overlays are displayed on graphics and movies
inTag registered:
-no limitations
-no overlays
-free (within a collaboration for academic institutions)
-we encourage you to send us some representative images you get on your system if you are unexperienced with cardiac tagging or if your images doesn’t look like the demo sets, so that we can give you some advices with the settings your tagging sequences to fit the minimal requirements of inTag for proper handling of your data.
-all features are enabled for testing purposes
-can only process the provided datasets
-«inTag DEMO» overlays are displayed on graphics and movies
inTag registered:
-no limitations
-no overlays
-free (within a collaboration for academic institutions)
-we encourage you to send us some representative images you get on your system if you are unexperienced with cardiac tagging or if your images doesn’t look like the demo sets, so that we can give you some advices with the settings your tagging sequences to fit the minimal requirements of inTag for proper handling of your data.
0 Comments
what kind of MR-tagging images can be processed with inTag?
23/11/09
To read images, inTag relies on the DICOM parser built-in in OsiriX. The default and recommanded one is DCMTK (Dicom Offis), but it can optionally use DCM framework or Papyrus Toolkit.
Once images are in the database, inTag can process and has been designed to handle all kind of tagging patterns:
-grid tagging
-line tagging with both direction on the same serie
-line tagging with each direction in a separate serie
It is set in the first step by clicking on the kind of tagging you have.
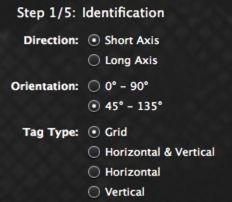
check the How-to section to see how to process different kind of images
It has been tested successfully with Siemens (product or WIP sequences) , Philips (product and WIP sequences) , GE, and Bruker MR tagged images, but you are welcome to send us your images to test them.
Once images are in the database, inTag can process and has been designed to handle all kind of tagging patterns:
-grid tagging
-line tagging with both direction on the same serie
-line tagging with each direction in a separate serie
It is set in the first step by clicking on the kind of tagging you have.

check the How-to section to see how to process different kind of images
It has been tested successfully with Siemens (product or WIP sequences) , Philips (product and WIP sequences) , GE, and Bruker MR tagged images, but you are welcome to send us your images to test them.